main functions¶
The core functions of the tomography package.
-
tomography_tutorial.functions.calculate_covariance_matrix(img_stack, outname, kernelsize=10, overwrite=False)[source]¶ compute the covariance matrix. If the target file already exists and
overwrite=Falsethis function acts as a simple file reader.Parameters: - img_stack (numpy.ndarray) – the normalized SLC image stack
- outname (str) – the name of the file to be written
- kernelsize (int) – the boxcar smoothing dimension
- overwrite (bool) – overwrite an existing file? Otherwise it is read from file and returned
Returns: the covariance matrix
Return type:
-
tomography_tutorial.functions.capon_beam_forming_inversion(covmatrix, kz_array, outname, height=70, overwrite=False)[source]¶ perform the capon beam forming inversion to create the final tomographic result. If the target file already exists and
overwrite=Falsethis function acts as a simple file reader.Parameters: - covmatrix (numpy.ndarray) – the covariance matrix
- kz_array (numpy.ndarray) – the wave number stack
- outname (str) – the name of the file to be written
- height (int) – the maximum inversion height
- overwrite (bool) – overwrite an existing file? Otherwise it is read from file and returned
Returns: the tomographic array
Return type:
-
tomography_tutorial.functions.read_data(input, outname, overwrite=False)[source]¶ read the raw input data into a numpy array and write the results
Parameters: Returns: an array in 2D (one file) or 3D (multiple files)
Return type:
-
tomography_tutorial.functions.start(notebook)[source]¶ Create a custom copy of the jupyter notebook with a name defined buy the user and start it. The notebook is only copied from the package if it does not yet exist. Jupyter notebook files have the extension ‘.ipynb’. If the defined notebook does not contain this extension it is appended automatically.
Parameters: directory (str) – the name of the custom notebook
-
tomography_tutorial.functions.topo_phase_removal(img_stack, dem_stack, outname, overwrite=False)[source]¶ Removal of Topographical Phase. If the target file already exists and
overwrite=Falsethis function acts as a simple file reader.Parameters: - img_stack (numpy.ndarray) – the SLC image stack
- dem_stack (numpy.ndarray) – the image stack containing flat earth and topographic phase
- outname (str) – the name of the file to be written
- overwrite (bool) – overwrite an existing file? Otherwise it is read from file and returned
Returns: the normalized SLC stack
Return type:
ancillary functions¶
Additional general functions for the tomography package.
-
tomography_tutorial.ancillary.cbfi(slice, nTrack, height)[source]¶ computation of capon beam forming inversion for a single pixel. This function is used internally by the core function
capon_beam_forming_inversion().Parameters: - slice (numpy.ndarray) – an array containing the covariance matrix and wave number for a single pixel
- nTrack (int) – the number of original SLC files
- height (int) – the maximum inversion height
Returns: the tomographic result for one pixel
Return type:
-
tomography_tutorial.ancillary.geocode(data, lut_rg_name, lut_az_name, outname=None, range_min=0, range_max=None, azimuth_min=0, azimuth_max=None)[source]¶ Geocode a radar image using lookup tables. The LUTs are expected to be georeferenced and contain range and azimuth radar coordinates for a specific image data set which is linked to these LUTs. If parameter data is a subset of this data set, the pixel coordinates of this subset need to be defined.
Parameters: - data (numpy.ndarray) – the image data in radar coordinates
- lut_rg_name (str) – the name of the range coordinates lookup table file
- lut_az_name (str) – the name of the azimuth coordinates lookup table file
- outname (str or None) – the name of the file to write;
if None, the geocoded array is returned and no file is written.
See function
geowrite()for details on how the file is written. - range_min (int) – the minimum range coordinate
- range_max (int) – the maximum range coordinate
- azimuth_min (int) – the minimum azimuth coordinate
- azimuth_max (int) – the maximum azimuth coordinate
Example
>>> from osgeo import gdal >>> from tomography_tutorial.ancillary import geocode >>> image_name = 'path/to/somedata/image.tif' >>> lut_rg_name = 'path/to/somedata/lut_rg.tif' >>> lut_az_name = 'path/to/somedata/lut_az.tif' >>> outname = 'path/to/somedata/image_sub_geo.tif' >>> image_ras = gdal.Open(image_name) >>> image_mat = image_ras.ReadAsArray() >>> image_ras = None >>> image_mat_sub = image_mat[0:100, 200:400] >>> geocode(image_mat_sub, outname, lut_rg_name, lut_az_name, range_min=200, range_max=400, azimuth_min=0, azimuth_max=100)
-
tomography_tutorial.ancillary.geowrite(data, outname, reference, indices, nodata=-99)[source]¶ write an array to a file using an already geocoded file as reference. The output format is either GeoTiff (for 2D arrays) or ENVI (for 3D arrays).
Parameters: - data (numpy.ndarray) – the array to write to the file; must be either 2D or 3D
- outname (str) – the file name
- reference (gdal.Dataset) – the geocoded reference dataset
- indices (tuple of slices) – the slices which define the subset of data in reference,
i.e. where is the data located within the reference pixel dimensions;
see
lut_crop() - nodata (int) – the nodata value to write to the file
-
tomography_tutorial.ancillary.listfiles(path, pattern)[source]¶ list files in a directory whose names match a regular expression
Parameters: Returns: a list of absolute file names
Return type: Example
>>> listfiles('/path/to/somedata', 'file[0-9].tif') ['/path/to/somedata/file1.tif', '/path/to/somedata/file2.tif', '/path/to/somedata/file3.tif']
-
tomography_tutorial.ancillary.lut_crop(lut_rg, lut_az, range_min=0, range_max=None, azimuth_min=0, azimuth_max=None)[source]¶ compute indices for subsetting the range and azimuth lookup tables (LUTs). The returned slices describe the minimum LUT subset, which contains all radar coordinates within the range-azimuth subset.
Parameters: - lut_rg (numpy.ndarray) – the lookup table for range direction
- lut_az (numpy.ndarray) – the lookup table for azimuth direction
- range_min (int) – first range pixel
- range_max (int) – last range pixel
- azimuth_min (int) – first azimuth pixel
- azimuth_max (int) – last azimuth pixel
Returns: the pixel indices for subsetting: (ymin:ymax, xmin:xmax)
Return type: tuple of slices
-
tomography_tutorial.ancillary.normalize(slice)[source]¶ normalize a 1D array by its minimum and maximum values:
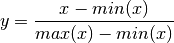
Parameters: slice (numpy.ndarray) – the 1d input array to be normalized Returns: the normalized array Return type: numpy.ndarray
plotting¶
Plotting utilities for the Jupyter notebook.
-
class
tomography_tutorial.plotting.DataViewer(slc_list, phase_list, kz_list, slc_stack, phase_stack, kz_stack)[source]¶ functionality for displaying the input data (SLC, topographic phase and wave number)
Parameters: - slc_list (list of str) – the names of the SLC input files
- phase_list (list of str) – the names of the topographic phase input files
- kz_list (list of str) – the names of the Kappa-Zeta wave number input files
- slc_stack (numpy.ndarray) – the SLC images
- phase_stack (numpy.ndarray) – the topographic phase images
- kz_stack (numpy.ndarray) – the wave number images
-
class
tomography_tutorial.plotting.GeoViewer(filename, cmap='jet', band_indices=None)[source]¶ plotting utility for displaying a geocoded image stack file.
On moving the slider, the band at the slider position is read from the file and displayed.
Parameters: - filename (str) – the name of the file to display
- cmap (str) – the color map for displaying the image. See
matplotlib.pyplot.imshow(). - band_indices (list) – a list of indices for renaming the individual bands in filename such that one can scroll trough the range of inversion heights, e.g. -70:70, instead of the raw band indices, e.g. 1:140. The number of unique elements must of same length as the number of bands in filename.
-
class
tomography_tutorial.plotting.Tomographyplot(capon_bf_abs, caponnorm)[source]¶ functionality for creating the main tomography_tutorial analysis plot
Parameters: - capon_bf_abs (numpy.ndarray) – the absolute result of the capon beam forming inversion
- caponnorm (numpy.ndarray) – the normalized version of capon_bf_abs;
see function
normalize().